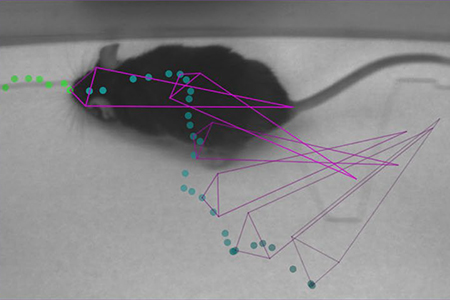
Photo courtesy of Dr. Alexander Mathis
Many of the commercial systems and open source software available can record and analyze complex body movement of animals in a variety of user-friendly interfaces with high throughput7-10,13. However, they are often rigid in their use, rely on expensive licenses, can require invasive markers, and/or have specific hardware requirements.
Researchers need systems which can precisely observe animal behavior, but with greater flexibility and reduced overhead requirements. This is the niche that machine learning systems, such as DeepLabCut, are attempting to fill.
Observing Animal Behavior with Machine Learning
Mathis and colleagues published their technical report in the September issue of Nature Neuroscience1. DeepLabCut is an open source Python toolbox for using user-labelled images to train tailored networks to perform automatic labeling of novel data2,3.DeepLabCut provides a robust and powerful machine learning platform (based on DeeperCut, pretrained on ImageNet) to predict the location of a body part without the need for markers, providing a flexible detection system1,4,5.
Challenges of Observing Animal Models
In current video analysis of hand movement detection, markers are often difficult to use and can inhibit full articulation of the joints1. Furthermore, in the real-world laboratory, use of video tracking introduces a multitude of confounding factors, such as changing light conditions, lens distortions, shadows, and animal body part distortions based on environmental interactions.Commercial systems and computational video analysis can address many of these in preprocessing steps, e.g. camera calibrations. Yet, it can be a tedious process and cannot account for all variables that can distort video detection during an experiment.
To test its ability to detect over a variable visual landscape, DeepLabCut was evaluated without the use of preprocessing1. The team wanted to test DeepLabCut's ability to:
- Locate and track body parts without predefined markers
- Evolve its ability to do so as its data set grows
- Function in a variety of behavioral and environmental setups
- Support maximum user flexibility and transparency
- Detect and observe behavior faster and more accurately
DeepLabCut Performance
DeepLabCut was able to approach human detection quality when tested after training1. They also varied the size of the training sets to compare the number of trained images with the reported error size. Thus, they were able to determine the number of training frames required for "excellent" generalization of a given body location indicative of a set behavior1.The algorithm is also highly data-efficient, in that, data augmentation alone does not provide for any further reduction in error. However, increasing the number of images that capture behavioral variability did produce a performance gain. Interestingly, DeepLabCut performed significantly better when trained on all body parts of a given animal compared to a more focused training approach, such as only observing a single part. This holds true even when both networks (all body parts vs. specific body part) have the same amount of data on the given specific body1.
DeepLabCut was able to provide excellent detection with a variety of training size sets to: odor detection (required snout, ear, and tail tracking) in a mouse; body tracking in freely behaving Drosophila (notable background and orientation challenges); and a skilled reaching task (pulling a joystick requiring individual digit tracking) of a mouse's hand1. The more complex a behavioral detection and/or setting, the more it increased the need for training image sets. However, DeepLabCut was able to detect highly enriched hand pose estimation in a mouse with only 141 training frames representing 5 different mice1.
Harnessing Pre-Trained Data Sets
A key difference in DeepLabCut compared to established detection platforms is its ability to demonstrate markerless generalization and transfer learning of a trained network1. The algorithm trained to only one mouse's body parts could recognize the body parts of novel mice with different body sizes. The task was not error-free but provided a case for body location detection trained in a single animal extending to multiples within the same image, which could be useful in social paradigms1.The power of DeepLabCut is in its ability to harness the full neural network of DeeperCut, which is used to learn and predict human body postures in a robust way4,5. Furthermore, the ability of DeepLabCut to demonstrate transfer learning (pretrained models to new tasks) can reduce the time and data necessary to accurately detect a given behavior in a laboratory setting.
Importance of Training Sets
DeepLabCut's highly-flexible algorithm allows for quick application to highly diverse behaviors with equally diverse video quality outputs, e.g. hand movements in a mouse vs. freely moving winged insects. It is a welcome addition to the animal detection/tracking platform. However, DeepLabCut is only as good as the training set volume, diversity, and accuracy of both labeling by the user and reflection of experimental (unlabeled) data.The flexibility of the system requires users to adequately define their training set and its diversity based on the behavior they want to analyze. The ability of the algorithm to generalize can also lead to suboptimal sampling if given a noisy or sparse behavior lacking adequate representation within the training set1. This can be addressed by the user in post hoc fine-tuning of the network, but requires knowledge of how the system arrives at its estimations.
Accessibility for Researchers
DeepLabCut is not a plug-and-play option. Users must be familiar with coding in Python (although a step by step guide is provided by Mathis and colleagues) and the visual requirements of the network to reliably capture their experimental behavior3. This is in contrast to many commercial systems that often have user-friendly interfaces, customer support, automated functions, and established, preprogrammed computational methods for key behavioral captures. The trade-off is often the rigidity and cost of the plug-and-play system.Summary
DeepLabCut provides an open source software tool that harnesses the power of the DeeperCut platform4,5. Investigators were able to show it accurately extracted image frames from video and could match human labeling accuracy after exposure to a sufficient training set. This trained network could then be used to label body part locations in unlabeled (novel) data with high accuracy.The plasticity of DeepLabCut can provide a profound tool in the plethora of available detection software to provide an ever-increasing toolbox for investigators.






.jpg)

.jpg)
.jpg)
.jpg)
.jpg)





.jpg)
.jpg)


.jpg)



.jpg)




.jpg)

.jpg)
.jpg)




