Identifying Microbial Changes in New-Onset Crohn's Disease
Multiple studies have shown that the intestinal microbiome is altered in Crohn's; however, it has been difficult to conclude whether these changes are causative, or merely a byproduct of chronic long-term inflammation.To address this problem, a recent study by Mottawea et al. focused on new-onset Crohn's disease in children1. Microbial alterations in these patients would more likely be causative and not affected by chronic inflammation or concomitant medications.
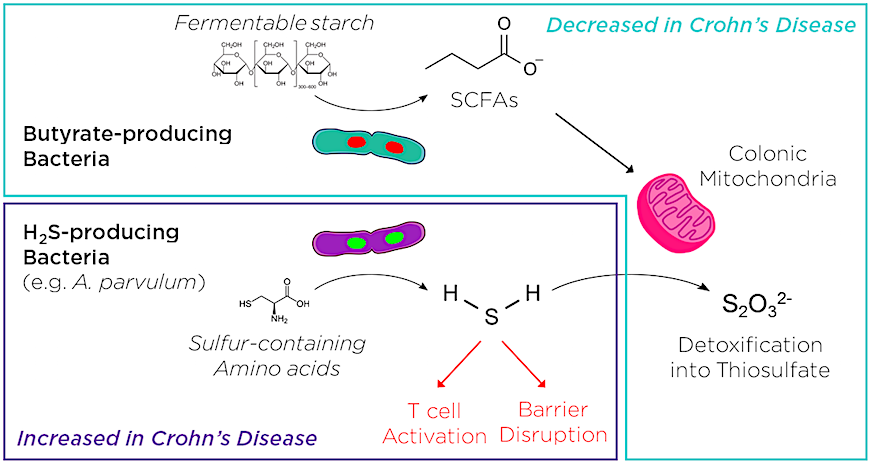
Like other studies using this approach,2,3 the authors identified a distinct microbial profile associated with disease. They found that Crohn's patients had a relative decrease in butyrate-producing bacteria and an increase in hydrogen sulfide (H2S)-producing bacteria. Butyrate, and other short chain fatty acids (SCFAs), are thought to be protective against inflammation. Excess H2S, which is normally detoxified through mitochondrial oxidation, may damage the intestinal barrier and contribute to T cell activation. This suggested that an imbalance in microbial butyrate/H2S production might contribute to Crohn's disease pathology.
Transkingdom Analysis Links Mitochondrial Function to A. Parvulum
Transkingdom correlation analysis is a novel approach which compares host variables (gene expression, proteomics, etc.) to microbial profiles to identify how specific taxa might influence disease. Mottawea et al. identified a reduction in mitochondrial H2S detoxification enzymes in biopsies from Crohn's patients, and used this approach to identify potential microbial culprits.Among the bacteria associated with reduced mitochondrial H2S detoxification, Atopobium parvulum stood out as a prominent node in the microbial network that defined the Crohn's disease microbiome. A. parvulum is an obligate anaerobic Gram positive cocci that is likely part of the normal human gut flora, but is also a known H2S producer.
It remained unclear how A. parvulum might influence mitochondrial function, or whether it was involved in Crohn's disease pathogenesis. The next logical step was to test this is a mouse model of colitis.
Connecting A. parvulum and Crohn's
To understand how A. parvulum contributes to disease, Mottawea et al. turned to a genetically engineered model of colitis, the interleukin 10 knockout mouse (IL10z-/-). IL10-/- mice spontaneously develop colitis over time and, importantly, this disease model requires an abnormal immune response to gut microbiota.When murine pathogen free IL10-/- mice were given A. parvulum, they quickly developed severe colitis. While this would suggest that A. parvulum is indeed pathogenic, an intriguing finding occurred when the authors tried the same experiment in the absence of commensal microbiota. When germ-free IL10-/- mice were mono-associated with A. parvulum, they did not develop any signs of disease.
The conclusion was that the pathogenic effect of A. parvulum required an interaction with other resident microbiota.
Microbial Networks in IBD Research
When researchers started looking at the IBD microbiome, there was an initial hope to identify specific pathogens - a microbial "smoking gun" - that caused the disease. This does not appear to be the case, and A. parvulum is now just one of a growing list of microbial factors associated with IBD.Understanding how networks of microbial communities vs. single pathogenic agents influence disease will involve a major leap from the traditional thinking lined out in Koch's postulates. The complex and dynamic relationship between the microbiome and health, involves a network of interactions between microbiota and the immune system4.
Modeling IBD on Germ-Free Mice
Mouse IBD models provide an ideal platform for investigating the influence of microbial networks on disease. Association studies in germ-free mice make it possible to investigate microbiomes at multiple levels:- Mono-association
- Simple defined flora (e.g. Altered Schaedler Flora, ASF)
- ASF in combination with specific microbes
- Increasingly complex (engineered) communities
- Mouse-derived microbiomes
- Patient-derived microbiomes
Conventional Mouse Models of IBD
Conventional mice remain an important tool in studying the mechanisms of IBD. The resident mouse microbiome is a major factor determining the severity of disease in IBD models. Controlling for variations between and within animal colonies is a major issue with regards to the reproducibility of mouse IBD studies.For instance, the presence or absence of specific commensals (e.g. Segmented Filamentous Bacteria, SFB) or pathobionts (e.g. Helicobacter spp., Klebsiella spp.) can influence the performance of IBD models, and should be considered in experimental designs.
Taconic offers mice at a variety of health standards, each of which excludes an increasingly more rigorous spectrum of microbial agents.















.jpg)

.jpg)
.jpg)
.jpg)
.jpg)





.jpg)


.jpg)
.jpg)

.jpg)


.jpg)





.jpg)

.jpg)





