Background: Types of Rodent Colitis Models
Colitis models are commonly grouped into three categories:- Chemically-induced colitis: Chemicals such as dextran sulfate sodium (DSS), 2,4,6-trinitrobenzene sulfonic acid or oxazolone are administered to standard strains, genetically engineered models (GEMs) or even humanized immune system mice to induce colitis.
- Adoptive transfer colitis: naïve T cells are harvested from a donor mouse and injected into an immunodeficient mouse of the same strain background to cause colitis. Commonly used donor/recipient pairs include BALB/c / C.B-17 scid and C57BL/6 / Rag2 knockout.
- GEM models of spontaneous colitis: Colitis occurs spontaneously due to gene knockout and response to the microbiome. Examples of GEMs that spontaneously develop colitis include Il10 and Mdr1a knockout mice.
IL-17 Blockade Ineffective for IBD
The promising strategy of IL-17 inhibition to treat IBD, derived from mouse model and clinical data, in fact failed in the clinic, with evidence that it actually worsened disease relative to placebo in patients with Crohn's disease2,3. While anti-IL-17A therapies can successfully treat the inflammatory diseases psoriasis and ankylosing spondylitis, there are now multiple case reports of IBD emergence in patients treated with the anti-IL-17A antibody secukinumab4,5,6.Mdr1a Knockout Mice Accurately Model Effect of IL-17 Inhibition on Colitis
Mice deficient for the p-glycoprotein efflux pump Mdr1a (Abcb1a) have defects in drug transport and are subject to spontaneous development of colitis. Colitis development is strongly influenced by the microbiome; disease can be exacerbated by infection with agents including specific bacteria, viruses and parasites7.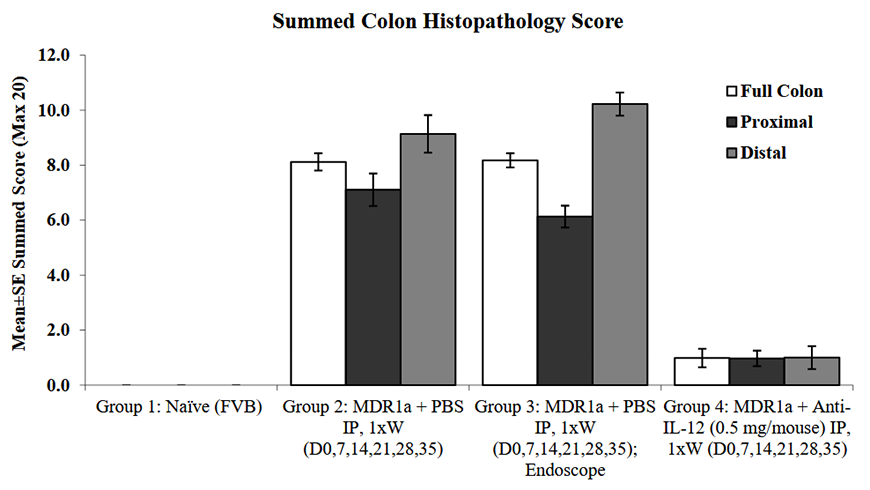
Figure 1: Mdr1a knockout mice spontaneously develop colitis, as demonstrated by comparison of summed histopathology score between control FVB mice and Mdr1a mice treated with either PBS (vehicle control). Anti-IL-12 treatment is efficacious at controlling colitis in Mdr1a mice. Data provided by Bolder BioPATH. Download the full data package, including protocol information, body weight change, endoscope scores and representative endoscopy images, disease activity scores, colon weights, histopathology scores and efficacy of anti-IL-12 treatment.
Treatment of Mdr1a knockout mice with an anti-mouse IL-17RA antibody rapidly exacerbated colitis, whereas in C57BL/6 mice administered DSS, IL-17RA inhibition had no effect on disease.Researchers at Amgen used the Mdr1a model to dissect the roles of individual IL-17 family members, identifying that "blockade of IL-17A specifically, but not IL-17E or IL-17F, leads to exacerbation of clinical signs of disease and mortality8."
This differential response of independent colitis models to a proposed therapeutic highlights underlines the importance of using multiple models and not relying on a single model to tell the full story.
Colitis Models as Mechanistic Tools Rather than Drug Screens
Preclinical colitis modeling is complex and it is not appropriate to draw conclusions about clinical efficacy from simple screens of drugs in a mouse model. However, these models are excellent tools for asking mechanistic questions, leading to a deeper understanding of IBD biology and pathways.As stated by Pizarro et al., "Every in vivo model has its strengths and weaknesses and the key to robust scientific investigation remains the selection of the correct model for the scientific question under inquiry with focus on mechanistic questions rather than direct translatability to efficacy in the clinic1."
The Future of Preclinical Colitis Modeling
Pizarro et al. note that more basic research is needed as "[e]ssential characterization of normal cellular and physiologic processes is lacking and is needed to advance the field". The authors suggest that models incorporating IBD patient-derived microbiota engrafted onto germ-free mice may be useful for research into therapies targeting dysbiosis.In addition to improvements in in vitro assays and data science, new and better validated animal models are needed. According to the review authors, "[t]hese could incorporate single or multiple genetic variants, humanized microbiota and/or humanized immune systems [and] could be used to recapitulate the gene-environment interactions that lead to the development of IBD, and be used to understand disease progression, triggers of disease flares, and prediction of disease trajectory and response to treatment1."















.jpg)

.jpg)
.jpg)
.jpg)
.jpg)





.jpg)


.jpg)
.jpg)

.jpg)


.jpg)





.jpg)

.jpg)




